Examples¶
The example section is divided into two subsections. In the first one, we infer subnetworks using in silico dataset with the aim to get familiar with Augusta. In the second section, we infer networks using a whole-genome organism´s dataset. All input data are available on GitHub in the “data” directory. Furthermore, in each section is a tutorial on how to download the inputs.
Test data¶
Test data are made by in silico dataset of bacterium´s Escherichia coli selected genes provided by the DREAM4 challenge.
Input files¶
count table file
We will use the file “insilico_size100_1_timeseries.csv” from the DREAM4 challenge.. We modified the file for our needs by swapping the matrix so that the genes are in the first column and times in the first row. We also rewrote the gene names according to the official gene names of the organism in order to match particular genes with information available in the GenBank file. The modified file is available on GitHub in the “data” directory as “Ecoli_DREAM4.csv”.
GenBank file
We will use Escherichia coli BW25113 available from the NCBI Nucleotide database under the accession CP009273. The GenBank file can be downloaded in the GenBank full format. Otherwise, it is also available on GitHub in the “data” directory as “Ecoli.gb”.
Usage example¶
Below is provided an example of the functions for generating GRN and converting GRN to BN. See Usage for further available functions.
Note: input files must be in the current directory or a full path must be provided.
Generate GRN:
>>> GRN = Augusta.RNASeq_to_GRN(count_table_input = 'Ecoli_DREAM4.csv', promoter_length = 1000, genbank_file_input = 'Ecoli.gb', normalization_type = 'TPM', motifs_max_time=180)[0]
Count table uploaded.
GenBank uploaded.
Count table normalization done.
Mutual information computation...
Mutual information computation done.
Motifs search...
Motifs search done.
Synonym organism names search...
Synonym genes names search...
Synonym names search done.
No data searched in interaction databases.
GRN stored as "GRN.csv".
The computation should be done in several minutes, depending on the specific machine.
Convert generated GRN to BN:
>>> Augusta.GRN_to_BN(GRN_input = GRN, promoter_length = 1000, genbank_file_input = 'Ecoli.gb', add_dbs_info = 1)
GenBank uploaded.
Synonym organism names search...
Synonym genes names search...
Synonym names search done.
No data searched in interaction databases.
GRN stored as "GRN.csv".
Cell Collective database search done.
Boolean network stored as "BN.sbml".
The computation should be done in several minutes, depending on the specific machine.
Refine GRN:
>>> refined_GRN = Augusta.refineGRN(GRN_input = GRN, genbank_file_input = 'Ecoli.gb', count_table_input = 'Ecoli_DREAM4.csv', promoter_length = 1000, motifs_max_time=180)
Count table uploaded.
GRN uploaded.
GenBank uploaded.
Motifs search...
Motifs search done.
Synonym organism names search...
Synonym genes names search...
Synonym names search done.
No data searched in interaction databases.
GRN stored as "GRN.csv".
The computation should be done in several minutes, depending on the specific machine.
Output files¶
The files are stored in the generated “output” directory.
motifs
all TFBM discovered in the genome assigned to their transcription factor
Stockholm file format
“discovered_motifs.sto”
available on GitHub in the “data/output” directory as “Ecoli_discovered_motifs.sto”
First transcription factor´s motifs in the file (transcription factor is BW25113_0564; discovered motif sequence is in the second column):
# STOCKHOLM 1.0
#=GF ID BW25113_0564
BW25113_0564_0 TTCTCCCCATCCTCCCAGGCATTACGCAACGTGAAACTCCAGGGATTTG
//
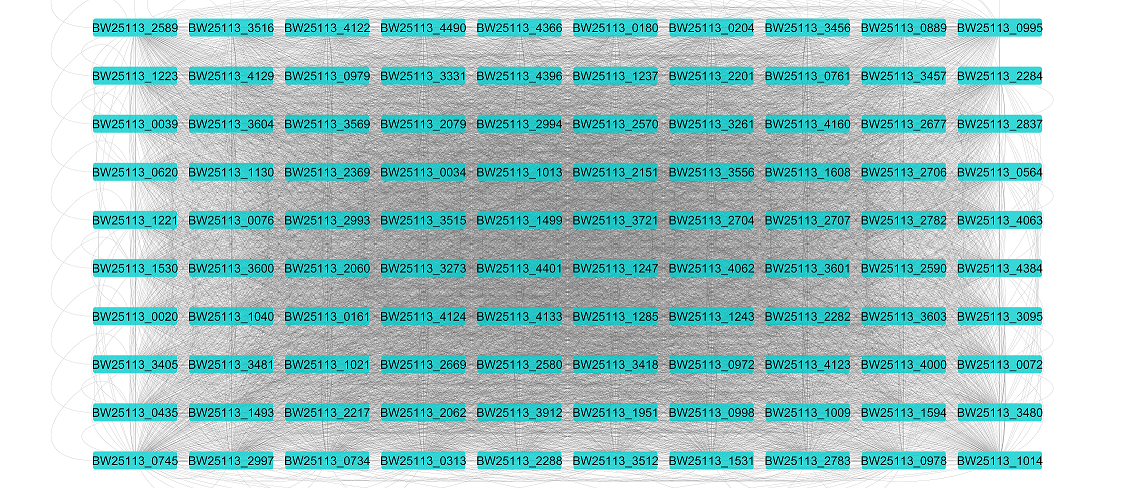
Gene Regulatory Network
adjacency matrix in the CSV file format
“GRN.csv”
available on GitHub in the “data/output” directory as “Ecoli_GRN.csv”.
GRN visualized in Cytoscape software:
Boolean Network
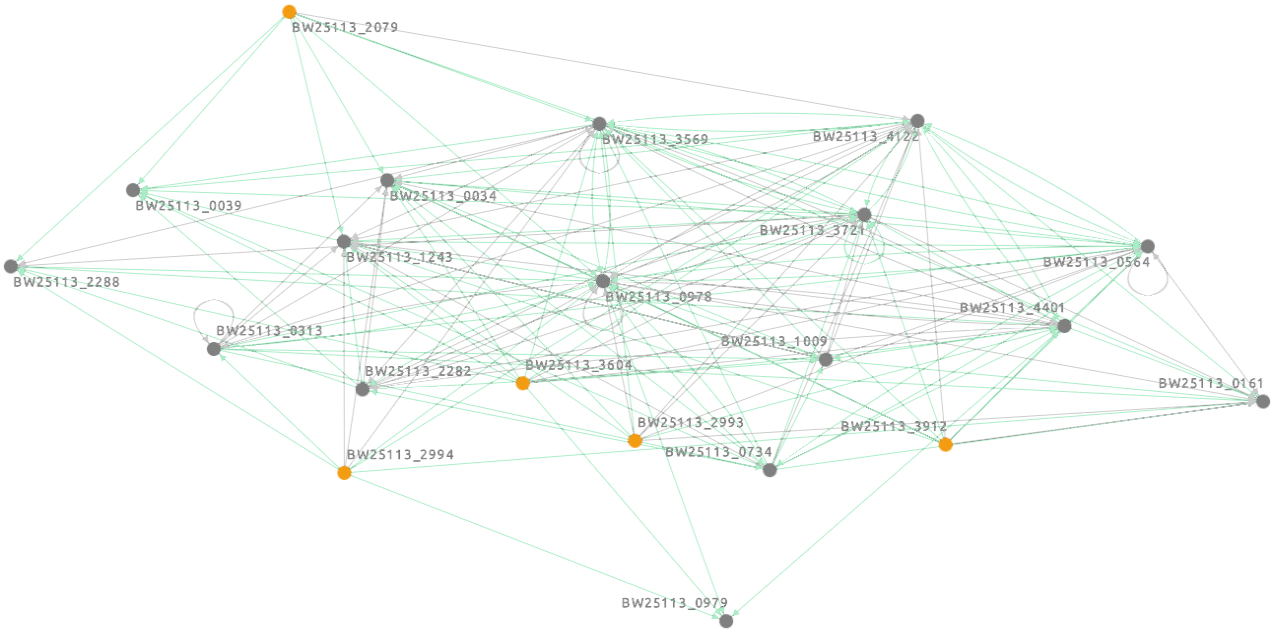
network in the SBML-qual file format
“BN.sbml”
available on GitHub in the “data/output” directory as “Ecoli_BN.sbml”.
BN (selected genes) visualized in Cell Collective platform:
Whole-genome data¶
The dataset consists of Clostridium beijerinckii NRL B-598 bacterium whole genome.
Input files¶
count table file
We will use the file we processed from the RNA-Seq dataset (available from the NCBI Sequence Read Archive (SRA) under the accession SRP033480; replicates B1 - B6). We generated and normalized the count table by R´s Rsubread and DESeq2 libraries. The processed count table is available on GitHub in the “data” directory as “Cbeijerinckii.csv”.
GenBank file
We will use C. beijerinckii NRL B-598 genome available from NCBI Nucleotide database under the accession CP011966.3. The GenBank file can be downloaded in the GenBank full format. Otherwise, it is also available on GitHub in the “data” directory as “Cbeijerinckii.gb”.
Usage example¶
Below is provided an example of the main function for generating a GRN and a BN. See Usage for further available functions.
Note: input files must be in the current directory or a full path must be provided.
>>> Augusta.RNASeq_to_BN(count_table_input = 'Cbeijerinckii.csv', promoter_length = 1000, genbank_file_input = 'Cbeijerinckii.gb', normalization_type = None, motifs_max_time = 300)
Count table uploaded.
GenBank uploaded.
Mutual information computation...
Mutual information computation done.
Motifs search...
Motifs search done.
Synonym organism names search...
Synonym genes names search...
Synonym names search done.
No data searched in interaction databases.
GRN stored as "GRN.csv".
Cell Collective database search done.
Boolean network stored as "BN.sbml".
The computation should be done in several days, depending on the specific machine.
Output files¶
The files are stored in the generated “output” directory.
transcription motifs as “discovered_motifs.sto”
Gene Regulatory Network as “GRN.csv”
Boolean Network as “BN.sbml”

